Autoinflammatory
diseases
Autoinflammatory diseases (AIDs) are rare, clinically and genetically heterogeneous diseases characterized by recurrent, self-limiting febrile episodes with systemic inflammation and involvement of serous membranes, synovial membranes and skin. The most serious complication is the risk of developing inflammatory amyloidosis, which can lead to end-stage renal disease. According to the pathophysiological pathways involved, AIDs are schematically divided into:
- inflammasomopathies (involving multiprotein complexes called inflammasomes)
- interferonopathies (associated with deregulation of type I interferon)
- other innate immune system impairments leading to inflammation
From a therapeutic point of view, depending on the gene and signaling pathway involved, specific biotherapies can be proposed to patients with AID (e.g., IL1 inhibitors are effective in some inflammasomopathies and JAK1/2 inhibitors in some type I interferonopathies).
The best known mendelian forms of AID are:
- Familial Mediterranean Fever (FMF), of autosomal recessive inheritance, linked to mutations in the MEFV gene (encoding pyrin)
- Cryopyrinopathies due to heterozygous mutations in NLRP3
- the autosomal dominant TRAPS (TNF-Receptor Associated Periodic Syndrome) syndrome related to mutations in TNFRSF1A,
- the autosomal recessive hyper-IgD syndrome caused by mutations in MVK
To date, more than 50 genes (such as NOD2, NLRP12, NLRC4, LPIN2, PSTPIP1, IL1RN, IL36RN, IL10RA, IL10RB, CARD14, PSMB8, TMEM173, TNFAIP3, TNFRS11A, and OTULIN) encoding key components of the innate immune system are involved in the pathophysiology of AID, with germline and somatic mosaic mutations identified from blood sampling.
Our team is interested in identifying the molecular etiologies of AID and understanding the underlying pathophysiological mechanisms, an essential step to improve diagnosis and management of patients. In the laboratory, we are studying
- the molecular aspect (panels of candidate ID genes, WES/WGS, whole exome/genome sequencing)
- the functional aspect in order to specify the signaling networks involved.
Depending on the candidate genes identified and the data that may be available on their function, we carry out functional studies in-vitro and ex-vivo. (see Figure 1).
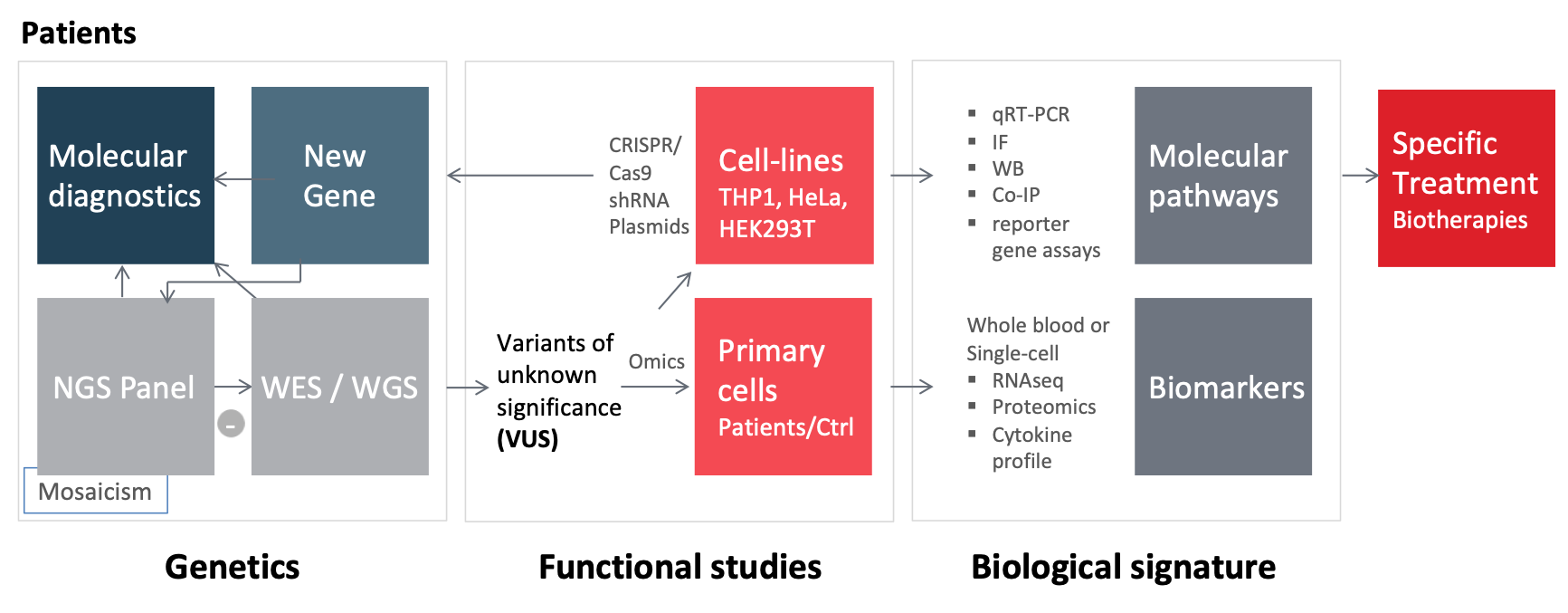
With this genetic and functional approach we have recently identified the p.Ala441Val missense variation in NLRP3 as a pathogenic recurrent mutational event in NLRP3-associated autoinflammatory disease1. We have also shown that in late-onset chronic urticaria, the search for autoinflammatory markers and somatic mosaic NLRP3 mutations may have important diagnostic and therapeutic consequences2. Moreover, our identification of mosaic mutations affecting the same NLRP3 amino acid (Glu569) led us to perform a comprehensive review of somatic and germline NLRP3 variants where we show that there are “hot spots” for somatic mutation, with only a few variants found in both the germline and somatic state. We conclude that the phenotypic spectrum of NLRP3-AIDs appears to be related to the germinal/mosaic status and localization of the underlying mutations3. Our work on inflammatory amyloidosis show that human monocytes and macrophages can express serum amyloid A genes, mainly SAA1 thus contributing to the local inflammatory microenvironment4.
References
1. Awad F, Assrawi E, Jumeau C, Odent S, Despert V, Cam G et al. The NLRP3 p.A441V Mutation in NLRP3-AID Pathogenesis: Functional Consequences, Phenotype-Genotype Correlations and Evidence for a Recurrent Mutational Event. ACR Open Rheumatol. 2019 Jun 6;1(4):267-276. doi: 10.1002/acr2.1039. PMID: 31777803.
2. Assrawi E, Louvrier C, Lepelletier C, Georgin-Lavialle S, Bouaziz JD et al. Somatic Mosaic NLRP3 Mutations and Inflammasome Activation in Late-Onset Chronic Urticaria. J Invest Dermatol. 2020 Apr;140(4):791-798.e2. doi: 10.1016/j.jid.2019.06.153. Epub 2019 Sep 9. PMID: 31513803.
3. Louvrier C, Assrawi E, El Khouri E, Melki I, Copin B et al. NLRP3-associated autoinflammatory diseases: Phenotypic and molecular characteristics of germline versus somatic mutations. J Allergy Clin Immunol. 2020 Apr;145(4):1254-1261. doi: 10.1016/j.jaci.2019.11.035. Epub 2019 Dec 6. PMID: 31816408.
4. Jumeau C, Awad F, Assrawi E, Cobret L, Duquesnoy P et al. Expression of SAA1, SAA2 and SAA4 genes in human primary monocytes and monocyte-derived macrophages. PLoS One. 2019 May 17;14(5):e0217005. doi: 10.1371/journal.pone.0217005. PMID: 31100086.
.
